Abstract: Researchers at Kanazawa University report in Proceedings of the National Academy of Sciences how a protein molecular motor can chop off a piece of cell membrane. The constrict-and-cut mechanism resembles that of a ratchet motor, and is of importance in processes mediating the entry of particles into cells.
Cells in the human body constantly receive substances from the outside via a process called endocytosis. One important endocytosis pathway involves the formation of a protrusion within the cell membrane, pointing to the inside of the cell, triggered when a molecule needing to get in reaches the membrane. The protrusion wraps and closes around the molecule, after which it is cut off, leaving the wrapped molecule (a so-called vesicle) inside the cell. A key role in the cutting-off mechanism is played by dynamin, a protein that can locally ‘constrict’ cell membranes, and chop off bits. The precise constriction mechanism is not completely understood, however. Now, by combining experiments and simulations probing the dynamics of dynamin, Alexander Mikhailov from Kanazawa University and colleagues show that the workings of such a ‘nanomuscle’ resembles that of a ratchet motor.
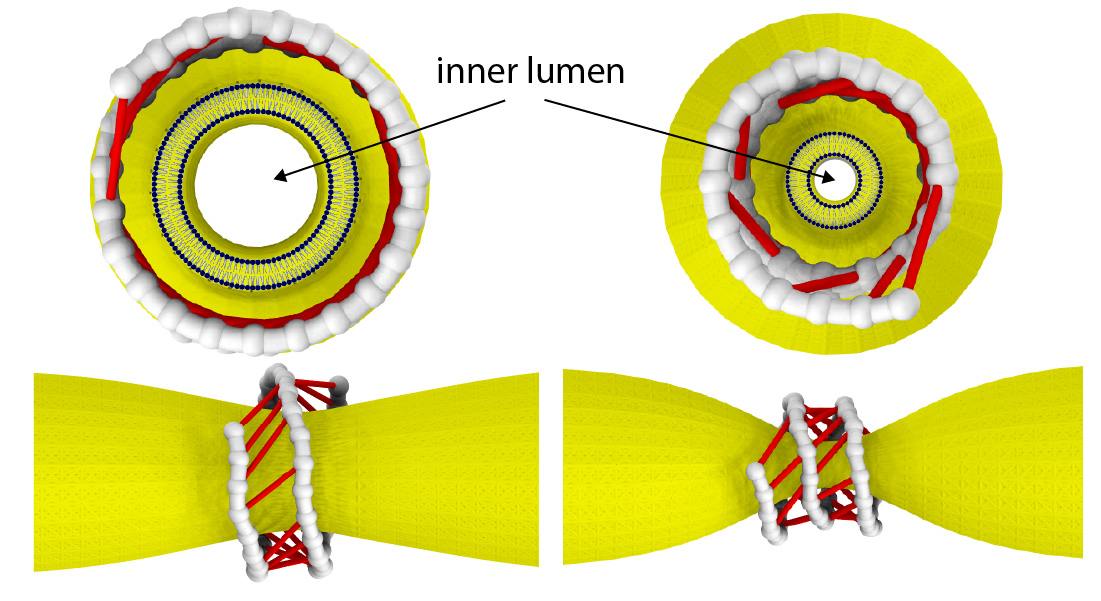
Dynamin consists of helical filaments, which coil around the neck of a vesicle forming within a membrane. In the presence of GTP (a molecule that can be thought of as a biochemical ‘fuel’, as it provides energy) these filaments start to slide, and in doing so constrict the vesicle neck. In order to gain detailed insights into the constriction mechanism, Mikhailov and colleagues used a technique called single molecule fluorescence resonance energy transfer (smFRET), which enables measuring distances in biomolecules. The obtained data revealed that different conformations of dynamin’s filaments occur, depending on whether or not they are bound to GTP and its products. These various configurations result in the formation of differently oriented molecular ‘bridges’ between neighboring coil turns. Supplying GTP and hydrolyzing it induces power strokes in the cross-bridges and generates a filament torque.
The scientists then designed a computable model of a set of dynamin filaments wrapped around a membrane protrusion, capturing the structural changes and their timescales as observed in the smFRET measurements. The membrane was modelled as a deformable cylindrical tube, and the filaments as strings of beads connected to active cross-bridges powered by GTP. The simulations showed that the combined motion of cross-bridges provides a force large enough to cut a membrane tube.
The work of Mikhalov and colleagues confirms, in a quantitative way, the constriction-by-ratchet model of the dynamin nanomuscle — nature’s strongest torque-generating motor. Quoting the researchers: “Our experimental and computational study provides an example of how collective motor action in megadalton molecular assemblies can be approached and explicitly resolved.”
Figure
Dynamin as a nanomuscle. The filament coils around a membrane tube and, under GTP hydrolysis, forms active cross-bridges that collectively constrict the membrane.
[Background]
Dynamin
Dynamin refers to a family of biomolecules. Classical dynamin is a protein playing a key role in endocytosis (the process through which particles are brought into a cell); the molecule is involved in the scission of vesicles formed from a cell membrane. Other molecules from the dynamin family are relevant for various biological processes including the division of organelles and cytokinesis (cell division).
Alexander Mikhailov from Kanazawa University and colleagues have now studied the scission of vesicles by dynamin in detail, and confirmed that when dynamin wraps around a tubular cell membrane fragment, it can constrict it, which results in scission.
smFRET
Single molecule fluorescence resonance energy transfer (smFRET) is a method used for measuring distances in the nanometer range in single molecules — typically biomolecules. smFRET is the application of FRET within a single molecule; FRET is the process of energy transfer between two chromophores (molecules that are sensitive to light). The efficiency of the energy transfer is inversely proportional to the sixth power of the distance between the chromophoric parts, making the FRET technique very sensitive to changes in distance.
Mikhailov and colleagues used smFRET to obtain information on the structure and conformational changes of dynamin, which was an important step in unraveling the constriction mechanism with which dynamin can cut off vesicles from a cell membrane.
[Article]
Title: Quantification and demonstration of the collective constriction-by-ratchet mechanism in the dynamin molecular motor
Journal: PNAS
Authors: Oleg M. Ganichkin, Renee Vancraenenbroeck, Gabriel Rosenblum, Hagen Hofmann, Alexander S. Mikhailov, Oliver Daumke, and Jeffrey K. Noel



 PAGE TOP
PAGE TOP