Abstract:Researchers at Kanazawa University examined a subset of GABA neurons in the circadian rhythm control center within the hypothalamus of the brain. They eliminated GABA signaling of vasopressin-producing neurons only in mice and found that it impaired circadian behavior. Specifically, time spent being active increased every day. Analysis showed a timing mismatch between the center’s molecular clock and the behavior. Thus, GABA signaling is required to make sure the timing remains in sync.
Our bodies and behaviors often seem to have rhythms of their own. Why do we go to the bathroom at the same time every day? Why do we feel off if we can’t go to sleep at the right time? Circadian rhythms are a behind-the-scenes force that shape many of our behaviors and our health. Michihiro Mieda and his team at Kanazawa University in Japan are researching how the brain’s circadian rhythm control center regulates behavior.
Termed the superchiasmatic nucleus, or SCN, the control center contains many types of neurons that transmit signals using the molecule GABA, but little is known about how each type contributes to our bodily rhythms. In their newest study, the researchers focused on GABA neurons that produce arginine vasopressin, a hormone that regulates kidney function and blood pressure in the body, and which the team recently showed is also involved in regulating the period of rhythms produced by the SCN in the brain.
To examine the function of these neurons, and only these neurons, the researchers first created mice in which a gene needed for GABA signaling between neurons was deleted only in vasopressin-producing SCN neurons. “We removed a gene that codes for a protein that allows GABA to be packaged before it is sent to other neurons,” explains Mieda. “Without packaging, none of the vasopressin neurons could send out any GABA signals.”
This means that these neurons could no longer communicate with the rest of the rhythm control center using GABA. On the surface, the results were simple. The mice showed longer periods of activity, beginning activity earlier and ending activity later than control mice. So, lack of the packaging gene in the neurons disrupted the molecular clock signal, right? In fact, the reality was not so simple. Closer examination showed that the molecular clock progresses correctly. So, what was happening?
The researchers used calcium imaging to examine the clock rhythms within the vasopressin neurons. They found that while the rhythm of activity matched the timing of behavior in control mice, this relationship was disturbed in the mice whose GABA transmission from the vasopressin neurons was missing. In contrast, the rhythm of SCN output, i.e. SCN neuronal electrical activity, in the modified mice had the same irregular rhythm as their behavior. “Our study shows that GABA signaling from vasopressin neurons in the suprachiasmatic nucleus help fix behavioral timing within the constraints of the molecular clock,” says Mieda.
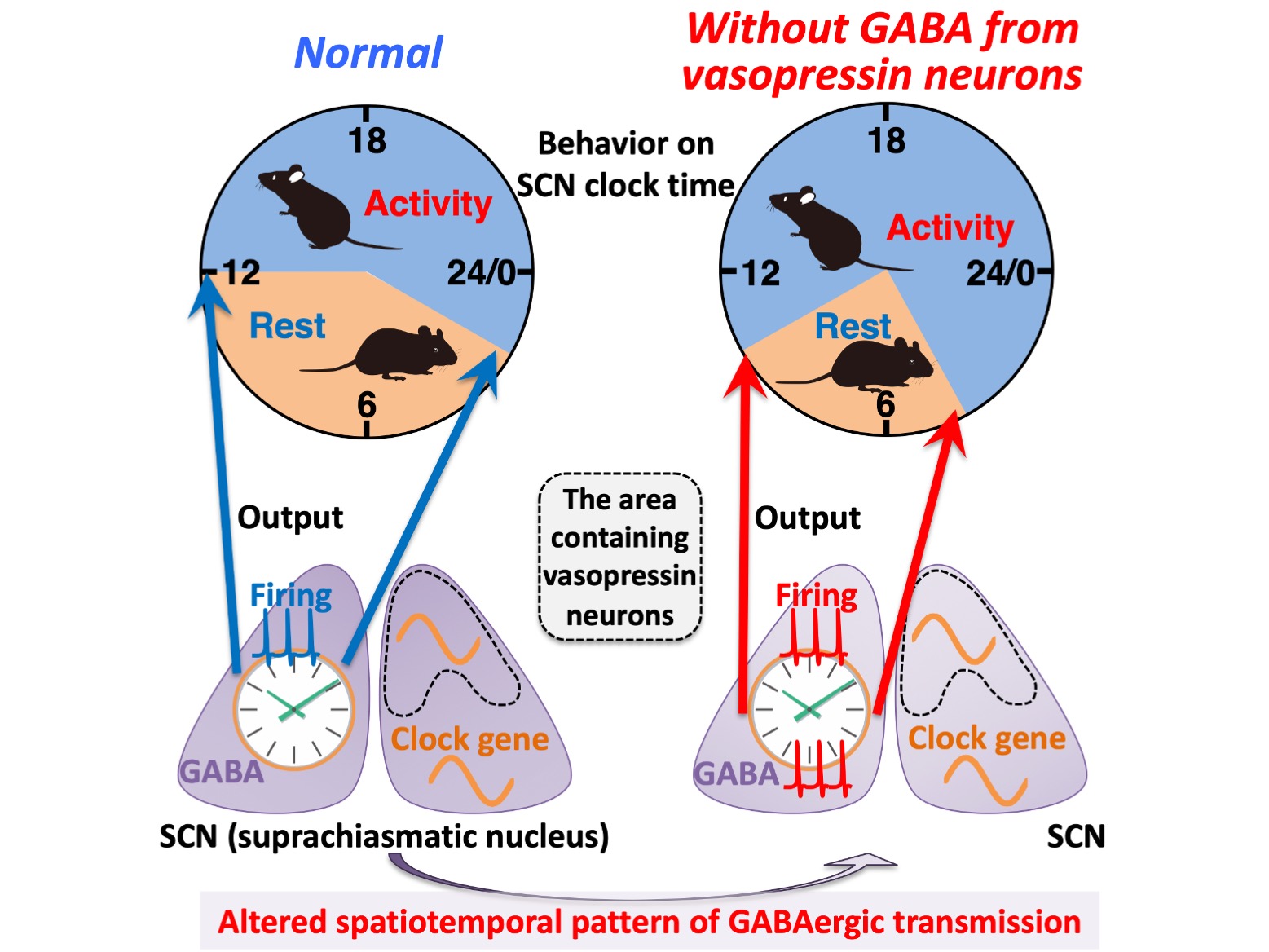
Figure.
A schema summarizing the effects caused by the deficiency of GABAergic transmission from vasopressin neurons on circadian rhythms at multiple levels. Without GABA release from vasopressin neurons, the spatiotemporal pattern of GABAergic transmission alters within the SCN. Such an alteration does not significantly disturb the spatiotemporal organization of molecular clocks measured with clock gene expression and intracellular calcium, but it does cause an aberrant bimodal pattern of the SCN firing (electrical activity) rhythm that may lead to the increased interval between the morning and evening locomotor activities. Thus, GABAergic transmission of vasopressin neurons regulates the SCN neuronal activity rhythm to modulate the time at which SCN molecular clocks enable circadian behavior.
[Article]
Title: GABA from vasopressin neurons regulates the time at which suprachiasmatic nucleus molecular clocks enable circadian behavior
Journal: PNAS
Authors:Takashi Maejima, Yusuke Tsuno, Shota Miyazaki, Yousuke Tsuneoka, Emi Hasegawa, Md Tarikul Islam, Ryosuke Enoki, Takahiro J. Nakamura, and Michihiro Mieda
[Funder]
This study was supported in part by Ministry of Education, Culture, Sports, Science, and Technology/Japan Society for the Promotion of Science (MEXT/JSPS) KAKENHI Grants 15K21745, 16H05120, 18H04941, 18H04972, 18K06519, 19H03399, 19K21241, 20K07259, 20K21498; The Japan Agency for Medical Research and Development Wise; the Takeda Science Foundation; the Uehara Memorial Foundation; by the Yamada Science Foundation; the Japan Foundation for Applied Enzymology; the Daiichi Sankyo Foundation of Life Science; the Kao Research Council for the Study of Healthcare Science; the Kanazawa University CHOZEN project.



 PAGE TOP
PAGE TOP