In a study recently published in Nature Genetics, researchers from Nano Life Science Institute (WPI-NanoLSI), Kanazawa University explore chromatin accessibility, i.e., endogenous access pathways to the genomic DNA, and its use as a tool for gene editing.
————————————————————————————————————————————————————————————————
Our DNA is protected from unwanted external modifications by forming structures called nucleosomes that consist of threads of DNA wound around chunks of special proteins known as histones. This unique coiled shape prevents the access of undesirable molecules to a cell’s DNA. However, for vital genetic functions—such as DNA repair—the right set of proteins require access to these DNA fragments. This phenomenon known as ‘chromatin accessibility’ involves a privileged set of protein molecules, many of which are still unknown.
Now, researchers from Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, led by Yusuke Miyanari, have used advanced genetic screening methods to unravel chromatin accessibility and its pathways.
For the investigation the team used a combination of two technologies—CRISPR screening and ATAC-see. While the former is a method to suppress the function of a desired set of genes, the latter is a means to identify which ones are essential for chromatin accessibility. Thus, using this method all genes playing a crucial role in chromatin accessibility could be pinned down.
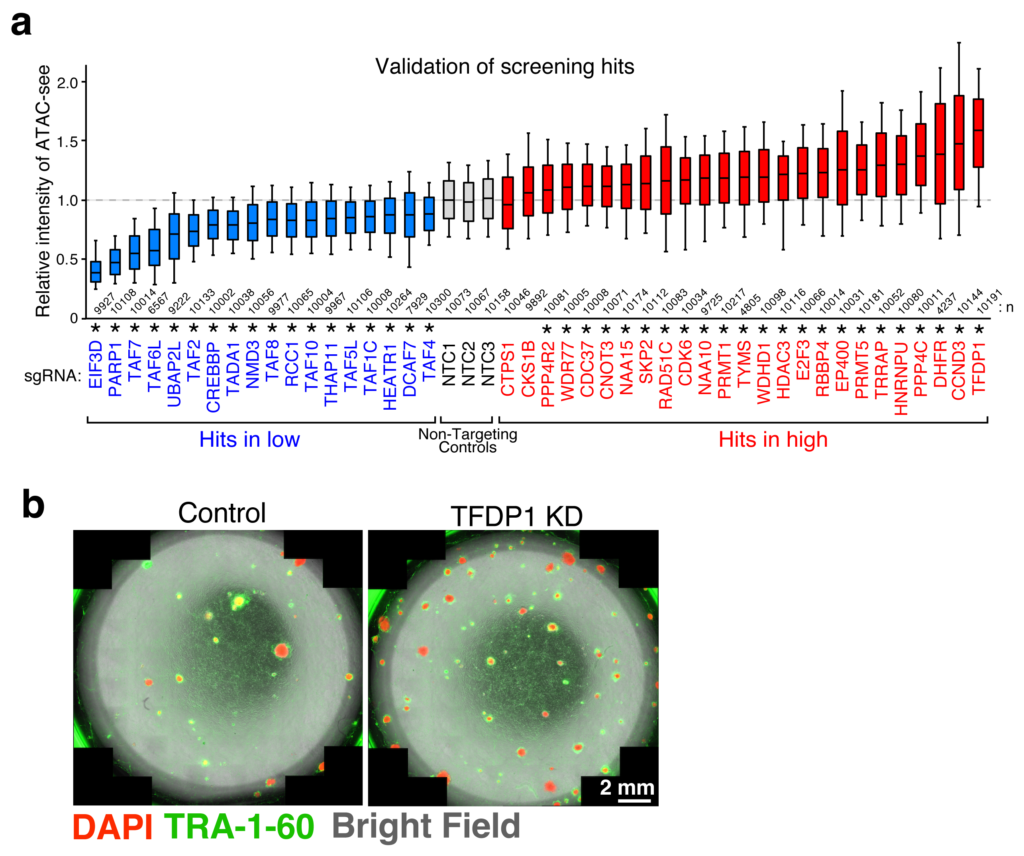
With the help of these assays, novel pathways and individual players involved in chromatin accessibility were uncovered—some playing a positive role and some negative. Of these, one particular protein, TFDP1, showed a negative effect on chromatin accessibility. When it was suppressed, a significant increase in chromatin accessibility was observed, accompanied by nucleosome reduction. A deeper dive into the mechanism of TFDP1 revealed that it functions by regulating the genes responsible for production of certain histone proteins.
The team then focused their study towards exploring biotechnological applications of their findings. After suppressing TFDP1, two different approaches were tried. The first approach involved gene editing using the CRISPR/Cas9 tool. This revealed that deletion of TFDP1 made the gene editing process easier. Now, most chromatin accessibility occurs in nucleosome-depleted regions or NDRs. However, by suppressing TFDP1 chromatin accessibility occurred not only in NDRs but across other regions as well. Secondly, the depletion of TFDP1 aided the process of converting regular cells into stem cells, a massive step forward in cellular transformation.
This study identified new chromatin accessibility pathways and channels for exploring its potential even further. “Our study shows the significant potential to manipulate chromatin accessibility as a novel strategy to enhance DNA-templated biological applications, including genome editing and cellular reprogramming,” conclude the researchers.
Fig 1:
a. Screening results of the players involved in chromatin accessibility analyzed by ATAC-see.
b. Successful reprogramming to stem cells (indicated by presence of TRA-1-60) after depletion of TFDP1 (right).
© 2024 Ishii, et al., under exclusive license to Springer Nature America, Inc.
Background
CRISPR screening
CRISPR screening is a large-scale tool used to find genes involved in a particular function. In this method, genes from the entire genome can be deleted one at a time to investigate which ones play a role in any target pathway. The mechanisms and roles of the genes narrowed down are then understood further using biological assays. In this study the genes identified by CRISPR screening were subjected to ATAC-see to confirm their involvement with chromatin accessibility.
ATAC-see
Transposase-Accessible Chromatin with visualization (ATAC-see) is a technique whereby accessible DNA locations can be visualized and quantified. ATAC-see involves using an enzyme to insert fluorescent proteins into the DNA at accessible sites. This technique works within nucleosome-depleted regions but also beyond these regions. Hence, when such sites were visualized after running CRISPR screening, the researchers could understand which genes were affecting chromatin accessibility directly.
Reference
Authors: Satoko Ishii, Taishi Kakizuka, Sung-Joon Park, Ayako Tagawa, Chiaki Sanbo, Hideyuki Tanabe, Yasuyuki Ohkawa, Mahito Nakanishi, Kenta Nakai, Yusuke Miyanari
Title: Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility
Journal: Nature Genetics
Published on Feb. 15, 2024.
DOI:10.1038/s41588-024-01658-1
URL: https://www.nature.com/articles/s41588-024-01658-1



 PAGE TOP
PAGE TOP