Researchers at Kanazawa University report in Biophysics and Physicobiology how to optimize high-speed atomic force microscopy experiments on live cell membranes, so that moving objects like molecules can be properly followed from frame to frame.
In video microscopy techniques, a practical issue is how to properly assign moving features. Indeed, any video recording setup has a finite frame rate, and if several identical (or similar) moving features appear in the frame, it is not trivial to understand which feature went where when comparing images recorded at successive time-points. For high-speed atomic force microscopy (HS-AFM), the assignment problem for highly dynamic samples — such as live cell membranes — has not been thoroughly investigated. Now, Damien Hall and Adam Foster from the NanoLSI at Kanazawa University have looked at feature assignment in HS-AFM from a theoretical point of view. They provide practical considerations for planning experiments on live cell membranes, in particular on how to find the optimal combination of frame size, image pixilation and sampling rate.
In AFM, an image is formed by scanning a surface with a very small tip with lateral dimension φ attached to a small cantilever. Horizontal (xy) scanning motion of the tip is controlled via piezoelectric elements; the vertical position of the tip changes as it follows the sample’s height profile, generating a force on the cantilever that can be measured and back-converted into a measure of the height, resulting in a height map of the sample’s surface. In HS-AFM, the surface is scanned repeatedly, resulting in a video.
The researchers first identified a set of key parameters that characterize HS-AFM experiments. Continuous spatial ‘sampling’ corresponds to the situation when the distance d between measured points (pixels) is smaller than the tip size, φ, whereas discontinuous sampling corresponds to d being larger than φ. Continuous sampling gives a better spatial resolution, but requires a larger scanning time τ — the time it takes to do a complete xy scan of the sample.
For mobile cell membranes, Hall and Foster distinguished between three confidence regimes for feature assignment, depending on how the scanning time τ compares with gross morphological changes in the membrane’s shape (typically of the order of minutes) and small-scale, rapid changes in the local composition of the membrane (of the order of milliseconds). If τ lies well above those two time scales, confidence is low. If it lies below both, confidence is high. If it lies in between, assignment confidence is intermediate. The idea is that for faster scanning (smaller τ), objects won’t have moved that much between frames and it is easier to recognize where objects have moved to.
The scientists considered the situation with no gross morphological changes — the intermediate- and high-confidence scenarios. Analytical and numerical descriptions of membrane diffusion, the xy scanning process and image pixilation made it possible, by invoking concepts from probability theory, to attribute ‘assignability’ properties to an image. Hall and Foster then developed criteria for confidently assigning single particles in an HS-AFM video, and also for the situation when multiple copies of one particles are present. An important result of their analysis is that how an image is pixilated plays a key role in feature assignability.
The work of Hall and Foster provides researchers with a checklist that should help them set up their HS-AFM experiment, taking into account basic setup parameters like scanning speed, minimum pixilation dimensions and scanned image size. Quoting the researchers: “We hope that the theoretical considerations developed here may prove useful to scientists using AFM in both the design and the interpretation of their experiments.” A software package called HS-AFM UGOKU is also available for download alongside the paper.
[Background]
High-speed atomic force microscopy
The general principle of atomic force microscopy (AFM) is to make a very small tip scan the surface of a sample. During this horizontal (xy) scan, the tip, which is attached to a small cantilever, follows the sample’s vertical (z) profile, inducing a force on the cantilever that can be measured. The magnitude of the force at the xy position can be related to the z value; the xyz data generated during a scan then result in a height map providing structural information about the investigated sample. In high-speed-AFM (HS-AFM), the working principle is slightly more involved: the cantilever is made to oscillate near its resonance frequency. When the tip is moved around a surface, the variations in the amplitude (or the frequency) of the cantilever’s oscillation — resulting from the tip’s interaction with the sample’s surface — are recorded, as these provide a measure for the local ‘z’ value. AFM does not involve lenses, so its resolution is not restricted by the so-called diffraction limit as in X-ray diffraction, for example.
HS-AFM results in a video, where the time interval between frames depends on the speed with which a single image can be generated (by xy-scanning the sample). Following moving objects from one frame to another is not trivial. Damien Hall and Adam Foster from Kanazawa University have now studied the ‘feature assignment problem’ for HS-AFM in detail, resulting in practical considerations for the optimization of HS-AFM experiments of live cell membranes.
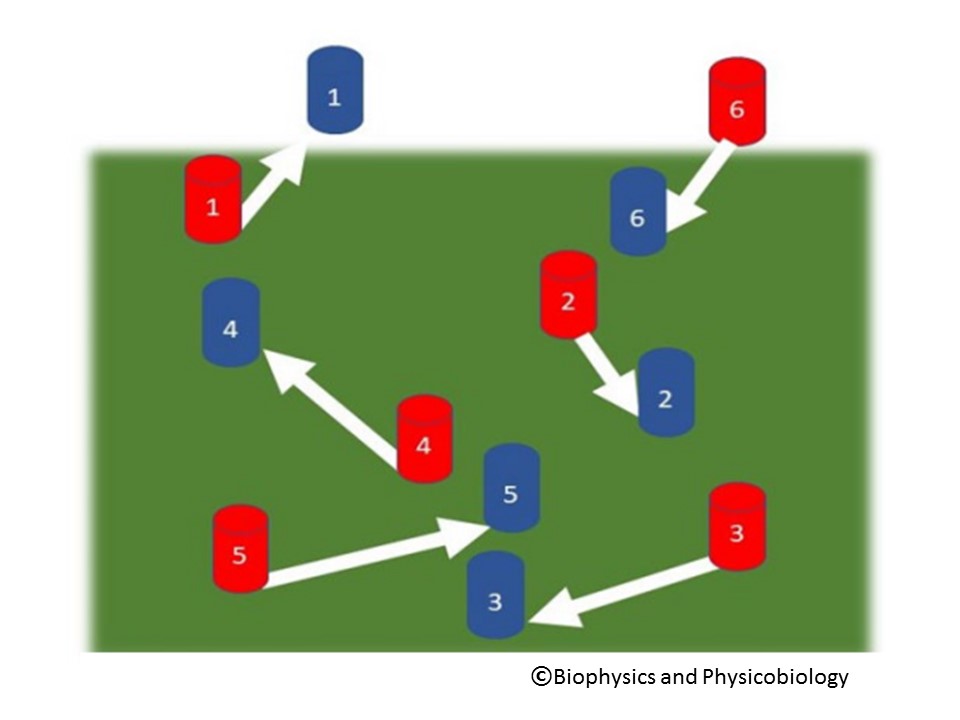
Figure. Feature assignment: procedures for determining how objects have moved between two successive video frames (red and blue) are not trivial.
License: CC BY-NC-SA
[HS-AFM UGOKU]
Software: https://jstagedata.jst.go.jp/articles/software/HS-AFM_UGOKU_User_Agreement/19609470
[Funder]
This work supported by KAKENHI Start-Up grant 21K20633, and the Academy of Finland under Project No. 314862.
[Reference]
Title: Practical considerations for feature assignment in high-speed AFM of live cell membranes
Journal: Biophysics and Physicobiology 19, e190016, 1-21 (2022).
Authors: Damien Hall, Adam S. Foster.
DOI: 10.2142/biophysico.bppb-v19.0016



 PAGE TOP
PAGE TOP